Kipoi Project - Sunset Announcement
After several impactful years, we have made the decision to archive the Kipoi repositories and end active maintenance of the project.
This is a bittersweet moment. While it’s always a little sad to sunset a project, the field of machine learning in genomics has evolved rapidly, with new technologies and platforms emerging that better meet current needs. Kipoi played an important role in its time, helping researchers share, reuse, and benchmark trained models in regulatory genomics. We’re proud of what it accomplished and grateful for the strong community support that made it possible.
Kipoi’s impact continues, however:
- The Kipoi webinar series will carry on, supporting discussions around model reuse and interpretability.
- Kipoiseq, our standard set of data-loaders for sequence-based modeling, also remains active and relevant.
Thanks to everyone who contributed, used, or supported Kipoi. It’s been a fantastic journey, and we're glad the project helped shape how models are shared in the field.
- The Kipoi Team

: Model zoo for genomics
Model zoo for genomics
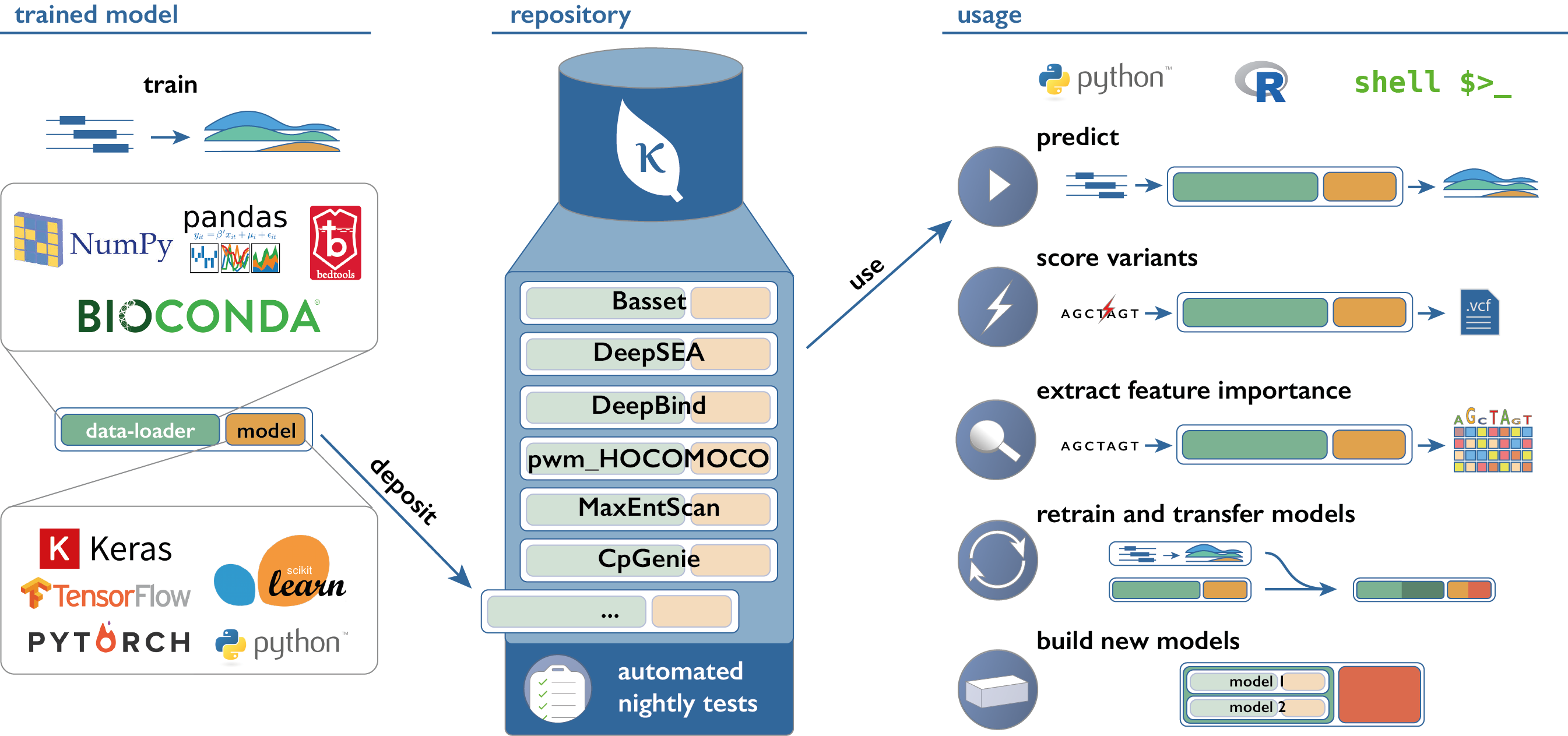
Kipoi (pronounce: kípi; from the Greek κήποι: gardens) is an API and a repository of ready-to-use trained models for genomics. It currently contains 2206 different models, covering canonical predictive tasks in transcriptional and post-transcriptional gene regulation. Kipoi's API is implemented as a python package (github.com/kipoi/kipoi) and it is also accessible from the command line or R.
Numbers
# of models: 2206
# of model groups: 38
# of contributors: 12
# of model groups supporting postprocessing:
- Variant effect prediction: 7/38
Model groups by framework
Model groups by tag
Getting Started
Use models
- Install Kipoi:
$ pip install kipoi - Choose the model from http://kipoi.org/models
- Use the model from CLI/python/bash by copying code snippets from http://kipoi.org/models/my_model
Contribute models
- Install Kipoi:
$ pip install kipoi - Run
$ kipoi ls - Explore the models locally at
~/.kipoi/models - Upload model weights to zenodo or figshare.
- Add the model to
~/.kipoi/models/(copy&edit the existing model or runkipoi init) - Test the model:
kipoi test ~/.kipoi/models/my_model - Submit the pull-request to github.com/kipoi/models
Contribute code
- Setup the development environment:
$ pip install kipoi[develop] - Read CONTRIBUTING.md
- Submit an issue/pull-request to github.com/kipoi/kipoi