Plugins
To enable additional functionality beyond just running model predictions, a plugin is available:
Kipoi-interpret
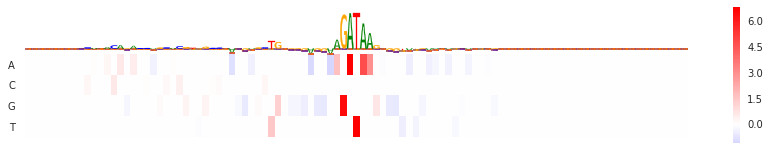
Kipoi-interpret is a general (genomics agnostic) plugin and allows to compute the feature importance scores like saliency maps or DeepLift for Kipoi models.
import kipoi
from kipoi_interpret.importance_scores.gradient import GradientXInput
model = kipoi.get_model("DeepBind/Homo_sapiens/TF/D00765.001_ChIP-seq_GATA1")
val = GradientXInput(model).score(seq_array)[0]
seqlogo_heatmap(val, val.T)